Phylogenomics Support Traditional Relationships within Cnidaria Zapata et al. 2015, PlosOne.11/9/2015 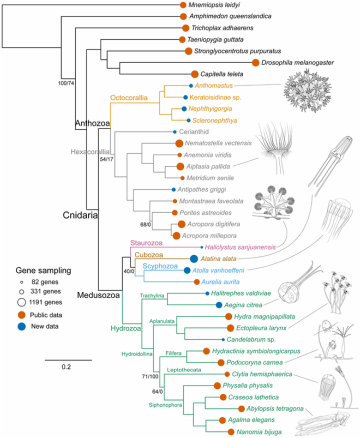 Figure 4, Zapata et al. 2015. PlosONE Figure 4, Zapata et al. 2015. PlosONE Felipe Zapata (postdoctoral researcher in the Dunn Lab) and colleagues used a transcriptomic approach to address uncertainties in evolutionary relationships within the Phylum Cnidaria. Their work was recently (Oct 14, 2015) published in PlosOne. This paper sets a milestone towards furthering our understanding of the evolution of a key group of basal metazoans with a broad range of life history traits, morphological characters, and ecological niches. Resolving evolutionary relationships among cnidarians (i.e., jellyfish, corals, anemones, hydroids) has proved challenging in part due to the ancient divergence (~500 million years) of the clade and the lack of single copy nuclear markers available for robust species tree analyses. Although the Medusozoa and Anthozoa have consistently been recovered as two monophyletic clades, alternative hypotheses have been suggested for the major lineages within each group. For example, long-standing views have recognized octocorals (sea fans, soft corals, sea pens) and hexacorals (stony corals, anemones) as sister taxa in the Anthozoa. This relationship was based on morphological characters, life cycle traits, and primarily on nuclear rDNA data. Recent studies, however, using mitochondrial genomes suggested that Anthozoa is paraphyletic, with octocorals sister to the medusozoans (Park et al. 2012; Kayal et al. 2013). Furthermore, Stampar et al. (2014) recovered the Order Ceriantharia (tube anemones) as sister to all remaining anthozoans, and proposed Ceriantharia as a new sub-class. With additional inconsistencies within the Medusozoa (Collins et al. 2006, Marques and Collins 2004), it has become clear that multiple, single-copy nuclear markers are sorely needed to resolve evolutionary relationships among cnidarians. Zapata and co-authors set out to address these uncertainties by using a phylogenomic approach to resolve relationships among major cnidarian lineages. They first sequenced transcriptomes of 15 new cnidarians, adding considerably to the existing genomic resources of cnidarians. Let me stress that again--Fifteen transcriptomes!! They then combined these newly sequenced data with existing transcriptomes for a dataset that included 31 cnidarians and 7 outgroups. [Three cnidarians had to be excluded from analysis due to poor (<5%) gene occupancy]. In total, they recovered 1,262 orthologous genes (365,159 aa), and generated a matrix with 38 taxa and 54% gene occupancy. Some people may argue that this is overall poor gene occupancy, but the majority of the nodes are in fact well supported. From my experience in working with a group (the Octocorallia) that has used only mitochondrial data and nuclear rDNA to construct phylogenies, I would say this work represents outstanding progress in Cnidaria Evolution. And, so what did Zapata et al. find? Their results strongly support several traditional views of Cnidaria, including monophyletic Anthozoa (octocorals and hexacorals) and Medusozoa clades. Also, these results further support the Marques and Collins (2004) cladistics work on Medusozoa, based on life history traits and morphology. Interestingly, this study also demonstrated the instability of the enigmatic Ceriantharia. This may be due to the low number of genes recovered for this group (16.6%) compared with others. It is clear that more data and more taxa are needed to resolve the relationship of Ceriantharia to other anthozoans. Zapata et al. (2015) has provided the scientific community with a wealth of new data to build upon and tools to use to further explore evolutionary hypotheses within the Cnidaria. I have to say these are exciting times for those working on the phylogenetics of any cnidarian group! AMQ Additional References: Collins et al. 2005 http://dx.doi.org/10.1080/10635150500433615 Kayal et al. 2013. http://www.biomedcentral.com/1471-2148/13 Marques and Collins 2004. http://onlinelibrary.wiley.com/doi/10.1111/j.1744-7410.2004.tb00139.x/abstract Park et al. 2012. http://www.sciencedirect.com/science/article/pii/S1055790311004374 Stampar et al. 2014 http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0086612
0 Comments
Leave a Reply. |
UCE Project TeamAll things Anthozoa, Evolution and Ecology Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation
Archives
September 2016
Categories |

 RSS Feed
RSS Feed
